Hypothetical Protein Rv1264/mt1302
(All numbering and residues are taken from first PDB file)
![]()
![]()
Bending Residue Dihedral Analysis
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
ILE-163
ALA-164
34.0
35.2
-5.3
12.3
34.4
33.5
-117.7
ALA-164
LYS-165
37.0
38.1
-0.4
-7.8
51.5
54.7
147.4
LYS-165
GLY-166
35.7
36.7
-10.9
4.7
62.5
67.6
107.7
GLY-166
SER-167
32.3
33.0
-2.6
-4.5
39.5
39.5
189.4
SER-167
GLN-168
33.9
33.7
-0.4
2.1
37.1
31.9
-112.7
GLN-168
ALA-169
35.8
36.0
-9.2
14.6
54.3
61.8
-57.1
ALA-169
LEU-170
32.8
33.6
-9.7
1.6
58.8
62.5
177.9
LEU-170
VAL-171
30.7
30.6
1.5
5.9
144.8
145.9
-136.7
VAL-171
SER-172
33.4
32.5
-8.2
14.3
46.7
36.8
-138.3
SER-172
GLN-173
34.2
33.8
4.0
19.9
122.0
121.5
-348.2
GLN-173
ILE-174
30.6
30.4
-0.9
24.8
69.8
65.9
-238.4
ILE-174
VAL-175
28.5
28.8
11.9
-8.5
127.6
147.5
-76.2
VAL-175
PRO-176
30.9
31.5
-1.9
1.8
10.9
21.5
76.4
PRO-176
LEU-177
31.2
30.6
-8.2
5.7
61.7
75.3
95.6
LEU-177
LEU-178
27.5
27.0
-7.7
-0.9
83.3
77.3
63.7
LEU-178
GLY-179
27.8
27.9
7.2
0.4
151.2
160.1
-124.7
GLY-179
PRO-180
29.5
29.0
-2.3
-4.2
47.0
60.6
45.9
PRO-180
MET-181
26.9
25.7
-2.7
7.3
104.5
109.3
53.0
MET-181
ILE-182
24.1
23.7
-4.7
12.9
76.0
71.3
-128.0
ILE-182
GLN-183
25.9
26.0
-14.1
0.6
9.4
19.1
201.1
GLN-183
ASP-184
26.2
25.5
-1.5
1.3
72.7
84.2
59.1
ASP-184
MET-185
22.5
21.7
0.1
-10.3
78.0
73.0
-11.2
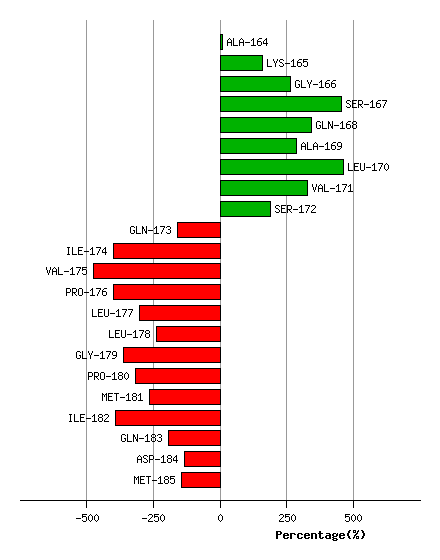
Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees