Fructose-1,6-Bisphosphatase (D-Fructose-1,6-Bisphosphate 3 1-Phosphohydrolase) (E.C.3.1.3.11)
(All numbering and residues are taken from first PDB file)
![]()
![]()
Bending Residue Dihedral Analysis
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
PRO-151
ASN-152
5.1
4.7
-13.5
-3.1
66.7
78.9
10.1
ASN-152
ASP-153
8.9
7.8
-9.4
36.7
11.0
15.0
-24.8
ASP-153
GLU-154
8.1
7.3
27.4
32.7
107.7
118.9
-16.6
GLU-154
CYS-155
4.9
7.1
-71.9
165.6
13.0
45.2
-68.2
CYS-155
ILE-156
3.2
4.2
-170.6
173.8
43.4
127.1
264.4
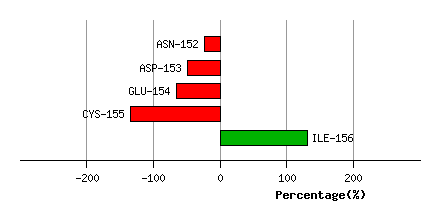
Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
ASN-177
VAL-178
2.1
2.1
169.3
7.3
133.0
157.4
170.0
VAL-178
CYS-179
2.9
3.4
157.9
47.2
148.5
136.4
-216.3
CYS-179
GLN-180
4.6
4.3
28.2
-92.5
129.5
106.6
41.4
GLN-180
PRO-181
3.0
2.9
-41.8
40.9
42.1
25.6
-7.5
PRO-181
GLY-182
5.4
5.6
-115.5
116.6
33.1
32.5
-3.8
GLY-182
ASP-183
4.0
4.6
-34.0
30.8
52.9
70.9
-2.0
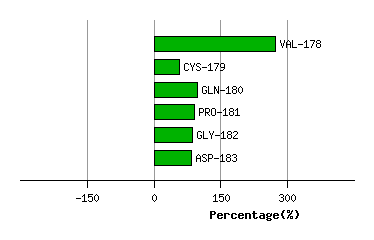
Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees