Transcription Antitermination Protein Nusg
(All numbering and residues are taken from first PDB file)
![]()
![]()
Bending Residue Dihedral Analysis
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
ASN-182
GLN-183
25.1
25.4
17.8
-8.9
57.2
68.3
55.5
GLN-183
ILE-184
22.5
22.8
17.8
-4.6
22.0
24.3
154.6
ILE-184
LYS-185
21.2
21.6
3.3
47.7
49.4
48.1
282.8
LYS-185
ARG-186
24.7
25.1
-19.1
-12.7
92.2
122.3
-88.2
ARG-186
GLY-187
26.9
25.2
86.3
-20.8
94.2
93.5
-75.1
GLY-187
VAL-188
24.7
21.9
-15.5
-59.1
145.3
134.4
-592.6
VAL-188
LYS-189
25.2
22.8
-101.0
-155.2
108.1
94.8
481.4
LYS-189
PRO-190
22.4
19.9
-168.5
-41.1
114.3
36.0
-838.5
PRO-190
SER-191
23.1
22.1
-171.2
-58.6
175.6
135.6
1181.9
SER-191
LYS-192
24.5
22.8
-73.1
13.4
116.0
136.1
-345.8
LYS-192
VAL-193
22.1
20.4
-165.8
104.6
44.4
65.0
301.6
VAL-193
GLU-194
19.7
19.0
-60.5
-32.7
65.9
126.4
-314.9
GLU-194
PHE-195
21.4
20.3
-179.7
-57.1
110.7
64.8
-7.3
PHE-195
GLU-196
20.4
20.0
-49.1
47.7
158.0
144.8
38.6
GLU-196
LYS-197
20.3
20.2
14.5
-25.5
85.2
94.9
-3.5
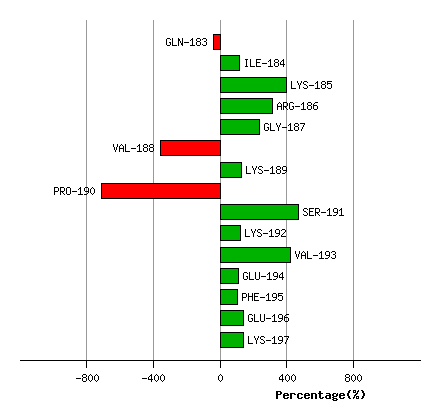
Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees