Skeletal Muscle Myosin II
(All numbering and residues are taken from first PDB file)
![]()
![]()
Bending Residue Dihedral Analysis
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
CYS-707
ARG-708
56.6
56.5
-0.1
0.0
138.6
138.4
0.0
ARG-708
LYS-709
56.8
56.8
0.1
-0.2
93.7
94.2
0.0
LYS-709
GLY-710
58.0
57.9
37.1
-29.3
110.3
110.2
-2.4
GLY-710
PHE-711
55.3
54.6
-0.1
0.0
126.4
131.5
0.0
PHE-711
PRO-712
51.7
51.1
0.0
0.1
100.9
92.6
-0.1
PRO-712
SER-713
51.3
50.7
-0.2
0.2
81.8
87.9
0.0
SER-713
ARG-714
47.7
47.1
0.0
-0.1
64.2
61.7
0.2
ARG-714
VAL-715
45.2
44.4
0.1
0.0
90.8
83.9
-0.1
VAL-715
LEU-716
41.5
40.8
0.0
-0.2
45.1
45.9
0.0
LEU-716
TYR-717
38.8
37.9
0.0
-0.1
55.4
54.6
0.0
TYR-717
ALA-718
36.2
35.5
0.2
0.0
65.2
57.7
-0.1
ALA-718
ASP-719
33.0
32.3
0.0
0.2
135.7
139.7
0.1
ASP-719
PHE-720
35.3
34.7
-0.1
-0.1
144.5
136.0
0.0
PHE-720
LYS-721
36.1
35.8
0.1
0.0
55.7
54.4
0.0
LYS-721
GLN-722
32.7
32.5
0.0
0.1
131.6
140.1
0.0
GLN-722
ARG-723
31.2
30.9
0.0
-0.1
32.4
34.5
0.0
ARG-723
TYR-724
34.1
33.9
0.2
0.0
43.0
51.0
0.0
TYR-724
ARG-725
34.5
34.7
0.1
-0.1
77.6
79.5
-0.1
ARG-725
VAL-726
31.3
31.7
0.0
0.0
166.9
162.6
0.0
VAL-726
LEU-727
30.8
31.4
0.1
0.2
54.6
63.0
0.0
LEU-727
ASN-728
33.9
34.8
-0.2
0.2
106.4
101.3
0.1
ASN-728
ALA-729
32.6
33.8
-0.1
0.0
60.4
65.5
-0.1
ALA-729
SER-730
29.4
30.4
0.1
0.1
121.0
122.2
0.0
SER-730
ALA-731
27.9
29.1
0.0
0.0
142.8
151.4
0.0
ALA-731
ILE-732
30.9
31.9
-0.1
0.1
47.2
49.1
0.0
ILE-732
PRO-733
32.0
32.6
-0.1
0.0
54.7
62.7
0.1
PRO-733
GLU-734
28.4
28.9
0.1
0.0
133.2
136.2
0.0
GLU-734
GLY-735
27.9
28.4
0.0
-0.1
140.8
146.9
0.0
GLY-735
GLN-736
31.2
31.4
0.0
0.0
132.7
125.7
0.0
GLN-736
PHE-737
30.5
30.3
0.1
0.0
125.5
119.4
0.0
PHE-737
MET-738
26.9
26.7
-0.1
0.1
49.0
43.3
0.0
MET-738
ASP-739
27.8
27.6
0.1
0.0
142.8
148.2
0.0
ASP-739
SER-740
30.5
29.9
0.1
0.0
80.1
78.1
0.0
SER-740
LYS-741
31.2
30.5
-0.1
0.1
103.4
97.4
0.1
LYS-741
LYS-742
34.8
34.1
0.1
0.0
48.1
55.3
-0.1
LYS-742
ALA-743
34.9
34.5
0.1
-0.1
17.6
9.0
0.1
ALA-743
SER-744
33.1
33.0
0.0
0.2
122.8
119.1
0.0
SER-744
GLU-745
35.9
35.8
-0.2
0.2
103.2
94.7
0.0
GLU-745
LYS-746
38.0
38.0
-0.1
0.2
149.8
146.9
0.1
LYS-746
LEU-747
36.0
36.4
-0.1
0.1
146.9
151.5
0.0
LEU-747
LEU-748
35.6
36.1
0.0
0.1
121.5
115.0
0.0
LEU-748
GLY-749
39.2
39.7
-0.1
0.0
120.2
113.0
-0.1
GLY-749
GLY-750
39.0
39.8
0.0
0.0
14.3
15.6
0.0
GLY-750
GLY-751
38.4
39.6
0.1
0.0
79.5
79.0
0.1
GLY-751
ASP-752
40.4
41.7
0.0
0.0
72.8
80.7
-0.1
ASP-752
VAL-753
43.6
45.1
0.1
-0.1
118.7
119.9
0.0
VAL-753
ASP-754
45.2
46.2
-0.1
0.1
56.9
48.3
0.0
ASP-754
HIS-755
48.4
49.2
-0.1
0.1
24.4
27.5
0.0
HIS-755
THR-756
47.1
47.7
-0.1
0.1
39.0
33.6
-0.1
THR-756
GLN-757
50.3
50.6
0.0
0.1
61.9
68.6
0.0
GLN-757
TYR-758
50.1
50.2
0.0
0.0
43.4
51.3
0.0
TYR-758
ALA-759
47.4
47.2
0.0
-0.1
38.1
31.4
0.1
ALA-759
PHE-760
46.8
46.4
0.0
0.0
53.4
61.2
0.0
PHE-760
GLY-761
43.9
43.2
0.1
-0.2
167.1
174.3
0.0
GLY-761
HIS-762
43.4
42.3
0.0
0.0
65.6
73.4
0.0
HIS-762
THR-763
40.8
39.4
0.0
0.1
100.1
108.6
0.0
VAL-765
PHE-766
43.0
42.4
0.0
0.0
95.6
94.8
0.0
PHE-766
PHE-767
46.4
45.8
0.1
-0.1
13.9
22.2
0.0
ALA-769
GLY-770
51.5
51.5
73.9
-120.0
27.9
27.3
24.7
GLY-770
LEU-771
49.8
50.7
-0.1
0.0
111.7
104.8
-0.1
LEU-771
LEU-772
47.0
47.4
0.0
0.1
97.4
114.2
0.0
LEU-772
GLY-773
44.4
45.7
0.0
0.0
125.8
115.5
0.1
GLY-773
LEU-774
45.8
47.4
0.1
-0.2
29.0
38.8
-0.1
LEU-774
LEU-775
44.8
45.3
0.0
0.1
87.5
73.8
0.0
LEU-775
GLU-776
41.3
42.2
-0.1
0.0
99.8
84.8
0.0
GLU-776
GLU-777
40.5
42.6
0.1
-0.1
21.0
35.1
0.0
GLU-777
MET-778
41.7
43.3
0.1
-0.1
57.3
67.4
-0.1
MET-778
ARG-779
39.1
39.9
0.1
0.0
108.3
125.0
0.1
ARG-779
ASP-780
36.7
38.6
0.0
-0.1
111.8
99.9
0.1
ASP-780
ASP-781
38.0
40.7
0.0
0.0
151.3
136.1
-0.1
LEU-783
ALA-784
32.8
36.3
-0.1
-0.1
134.4
121.3
0.0
ALA-784
GLU-785
34.0
37.7
0.1
-0.2
64.1
74.8
0.0
GLU-785
ILE-786
30.9
34.1
13.0
-2.3
123.9
140.3
-29.8
ILE-786
ILE-787
28.8
32.9
-18.9
25.1
96.6
82.9
-9.0
ILE-787
THR-788
27.6
31.0
0.1
0.1
29.7
42.5
0.1
THR-788
ALA-789
27.1
29.8
0.0
-0.1
99.9
87.0
-0.1
ALA-789
THR-790
25.3
28.4
-0.1
0.0
53.6
42.8
0.1
THR-790
GLN-791
23.2
26.9
0.1
-0.1
73.3
76.0
0.0
GLN-791
ALA-792
22.5
25.4
0.2
-0.2
50.2
63.9
-0.1
ALA-792
ARG-793
21.0
23.5
0.2
-0.1
104.8
118.5
-0.1
ARG-793
CYS-794
18.4
21.7
-0.1
0.0
71.2
67.3
0.1
CYS-794
ARG-795
17.3
20.5
0.1
0.0
41.2
46.6
0.0
ARG-795
GLY-796
17.5
19.7
-0.2
0.2
117.3
103.8
0.0
GLY-796
PHE-797
15.4
17.8
-0.1
0.0
59.5
47.6
0.1
PHE-797
LEU-798
13.0
16.2
-0.1
-0.1
84.9
83.8
0.0
LEU-798
MET-799
13.1
15.5
0.1
0.0
45.6
56.7
-0.1
MET-799
ARG-800
11.7
13.2
-0.1
0.0
92.1
79.4
0.0
ARG-800
VAL-801
9.2
11.9
0.0
0.0
60.0
50.0
0.0
VAL-801
GLU-802
7.4
9.9
-0.1
0.2
118.7
115.2
0.0
GLU-802
TYR-803
8.5
9.1
-0.2
0.2
132.1
118.4
0.0
TYR-803
ARG-804
5.9
6.9
-0.1
0.1
62.4
48.8
0.0
ARG-804
ALA-805
3.1
5.2
-0.1
-0.1
77.5
74.2
0.0
ALA-805
MET-806
5.8
4.8
-54.2
72.0
149.2
141.7
106.0
MET-806
VAL-807
5.3
4.5
-140.6
147.7
98.0
93.5
9.8
VAL-807
GLU-808
1.8
1.7
0.0
0.1
136.6
136.6
0.1
GLU-808
ARG-809
1.0
1.1
-0.1
0.1
103.2
103.3
-0.1
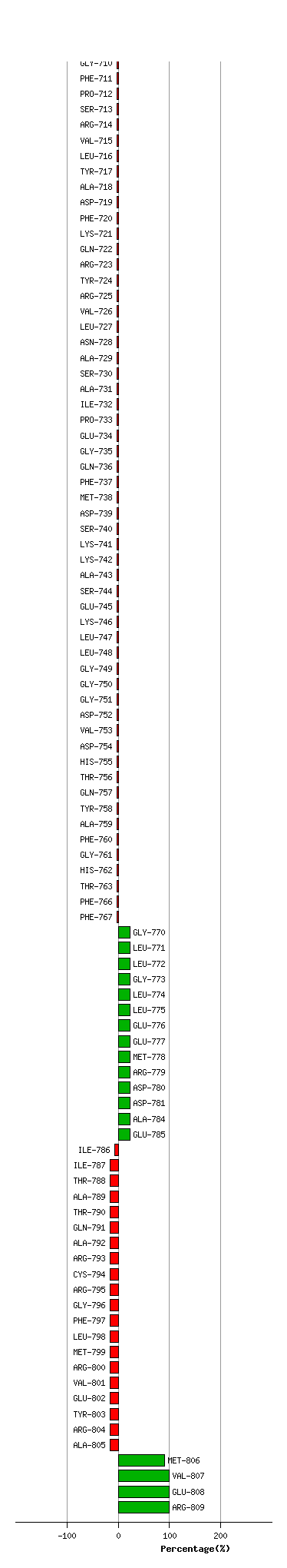
Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees