DNA Polymerase Iota
(All numbering and residues are taken from first PDB file)
![]()
![]()
Bending Residue Dihedral Analysis
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
SER-303
GLU-304
3.6
4.2
24.6
-57.7
18.8
33.5
-193.2
GLU-304
GLU-305
2.9
3.2
-77.0
73.8
104.5
111.8
-9.0
GLU-305
ASP-306
0.6
0.1
28.2
-15.0
36.6
46.8
115.4
ASP-306
SER-307
3.2
2.7
27.7
21.5
72.5
53.7
197.2
SER-307
PHE-308
4.8
4.3
91.2
-156.3
53.8
99.3
-173.3
PHE-308
LYS-309
7.7
8.0
-44.8
-84.5
84.2
45.0
226.6
LYS-309
LYS-310
10.3
9.2
-121.0
166.1
104.2
104.4
227.7
LYS-310
CYS-311
13.7
8.7
-29.8
-34.1
76.0
156.2
-218.7
CYS-311
SER-312
13.1
9.5
-29.9
-71.1
25.3
88.2
347.0
SER-312
SER-313
13.7
11.7
34.9
-91.7
100.9
133.1
-1279.9
SER-313
GLU-314
17.5
12.7
-120.2
85.0
29.0
83.2
-49.1
GLU-314
VAL-315
17.5
15.9
-4.5
-108.3
53.1
11.3
648.6
VAL-315
GLU-316
18.0
15.7
64.8
5.0
58.6
114.2
128.4
GLU-316
ALA-317
18.0
14.2
172.8
-27.2
89.0
164.0
-488.0
ALA-317
LYS-318
14.2
13.5
-158.2
98.3
20.2
77.4
347.2
LYS-318
ASN-319
13.2
14.7
4.3
-55.0
75.9
56.5
-120.1
ASN-319
LYS-320
14.0
12.4
7.5
-14.3
87.4
103.1
54.3
LYS-320
ILE-321
10.4
9.3
58.1
-74.7
142.0
151.3
88.1
ILE-321
GLU-322
9.3
9.4
0.0
-16.9
75.0
97.9
22.8
GLU-322
GLU-323
11.5
11.5
13.7
-33.0
56.1
43.5
-85.5
GLU-323
LEU-324
10.3
9.3
-3.0
-6.3
69.9
68.5
30.1
LEU-324
LEU-325
6.9
5.8
14.0
-50.5
141.0
131.5
182.0
LEU-325
ALA-326
8.7
7.7
77.8
-14.3
77.3
73.3
44.6
ALA-326
SER-327
11.7
10.6
-43.5
-6.0
95.1
72.2
60.5
SER-327
LEU-328
10.5
11.9
38.8
-7.7
145.6
137.7
-181.8
LEU-328
LEU-329
8.4
9.2
34.6
-52.0
135.1
151.1
89.8
LEU-329
ASN-330
11.2
11.7
-25.5
7.5
96.3
93.7
1.2
ASN-330
ARG-331
13.7
15.3
29.7
-49.1
96.2
76.4
-33.4
ARG-331
VAL-332
12.2
14.5
95.7
-68.4
160.6
127.9
-70.3
VAL-332
CYS-333
10.9
13.2
30.5
-45.6
121.8
134.5
3.8
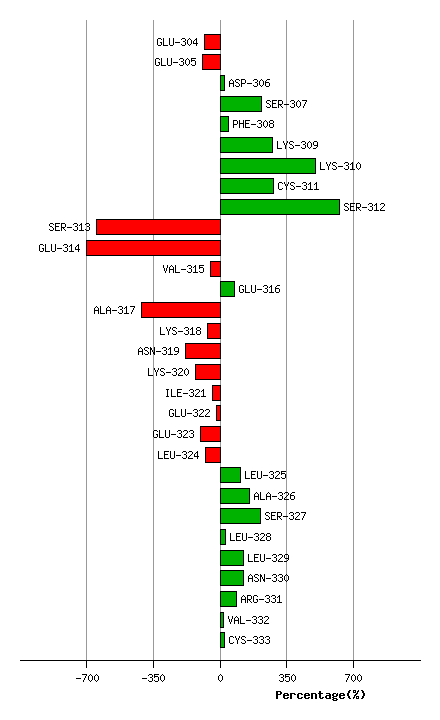
Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
PRO-339
HIS-340
7.1
7.8
17.7
-34.5
136.6
139.5
58.4
HIS-340
THR-341
4.7
5.0
20.1
-29.0
75.5
78.7
-3.0
THR-341
VAL-342
1.5
1.7
-9.0
11.0
156.8
165.2
26.1
VAL-342
ARG-343
2.0
1.4
0.5
-12.0
75.3
79.3
-34.8
ARG-343
LEU-344
2.9
2.8
-19.3
15.7
150.8
161.7
-27.3
LEU-344
ILE-345
5.5
5.0
-23.2
42.8
115.2
106.9
-1.7
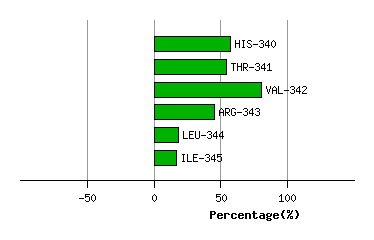
Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
GLU-358
SER-359
11.7
12.1
-58.6
57.4
111.3
103.2
23.8
SER-359
ARG-360
9.0
8.7
41.2
-42.5
20.2
41.0
-4.3
ARG-360
GLN-361
8.5
7.6
-13.4
-9.2
108.5
111.2
-88.2
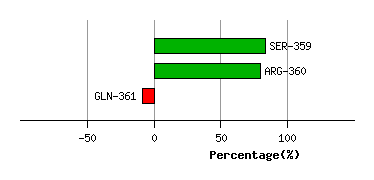
Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
MET-388
LYS-389
13.1
12.9
93.8
-96.1
121.8
139.9
22.4
LYS-389
LEU-390
13.7
13.5
4.2
-23.6
82.9
90.0
16.3
LEU-390
PHE-391
10.7
11.1
32.8
-35.8
32.6
27.3
4.3
PHE-391
ARG-392
12.1
12.5
-12.3
6.6
82.2
88.7
0.6
ARG-392
ASN-393
15.6
16.1
52.2
-69.3
114.4
126.4
74.4
ASN-393
MET-394
15.2
15.9
19.1
6.6
41.2
61.6
74.4
THR-404
LEU-405
11.2
10.8
139.2
-10.3
104.9
162.6
-397.6
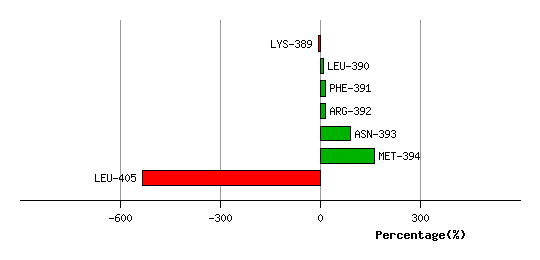
Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
SER-407
VAL-408
3.1
3.3
-42.8
12.1
163.0
161.1
-196.8
VAL-408
CYS-409
2.3
1.5
14.2
-4.5
77.5
73.4
-26.7
CYS-409
PHE-410
1.2
1.6
35.1
-33.7
24.8
39.2
38.6
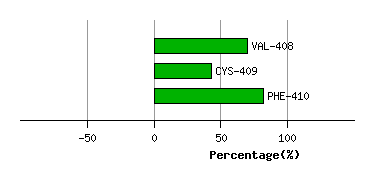
Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees