Fusion Protein Beta-2 Adrenergic Receptor/lysozyme
(All numbering and residues are taken from first PDB file)
![]()
![]()
Bending Residue Dihedral Analysis
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
VAL-222
PHE-223
4.5
4.1
1.0
1.6
139.1
120.0
2.3
PHE-223
GLN-224
6.4
5.5
-1.3
16.8
45.6
45.9
-10.1
GLN-224
GLU-225
3.6
2.9
8.7
-4.1
77.9
65.2
0.3
GLU-225
ALA-226
0.9
0.8
-24.7
16.1
93.7
104.8
-0.4
ALA-226
LYS-227
4.7
4.2
-9.4
65.2
25.7
37.2
-34.9
LEU-266
LYS-267
17.6
4.6
45.0
-7.0
24.7
56.1
-211.4
LYS-267
GLU-268
15.9
7.1
9.5
0.0
93.5
108.3
-3.4
GLU-268
HIS-269
12.7
9.7
162.6
-79.7
91.2
125.0
207.4
HIS-269
LYS-270
15.0
12.5
-123.4
1.2
149.0
99.3
-44.5
LYS-270
ALA-271
16.9
16.3
-14.5
28.9
132.8
159.3
9.2
ALA-271
LEU-272
13.4
16.6
-19.3
17.9
67.2
113.1
-2.0
LEU-272
LYS-273
12.4
14.6
-20.8
9.3
91.8
63.5
-6.6
LYS-273
THR-274
16.1
16.7
-12.0
-20.3
146.4
115.5
-19.9
THR-274
LEU-275
14.8
20.0
33.5
-4.2
93.3
13.2
11.9
LEU-275
GLY-276
12.6
18.7
-21.8
0.3
59.3
92.7
-1.8
GLY-276
ILE-277
15.7
18.7
-4.9
-10.3
129.7
72.7
-5.1
ILE-277
ILE-278
18.1
21.9
13.9
-54.2
59.5
58.3
-32.5
ILE-278
MET-279
15.8
24.0
31.0
159.5
114.3
6.6
61.0
MET-279
GLY-280
16.1
22.3
171.8
-34.3
106.8
67.0
-169.9
GLY-280
THR-281
19.4
21.3
-167.6
-66.4
132.9
155.5
91.0
THR-281
PHE-282
21.6
23.7
152.7
-11.8
67.2
62.9
-40.0
PHE-282
THR-283
20.5
22.9
29.7
-46.0
126.5
59.2
9.1
THR-283
LEU-284
21.9
26.3
-161.8
-2.3
85.8
52.1
99.8
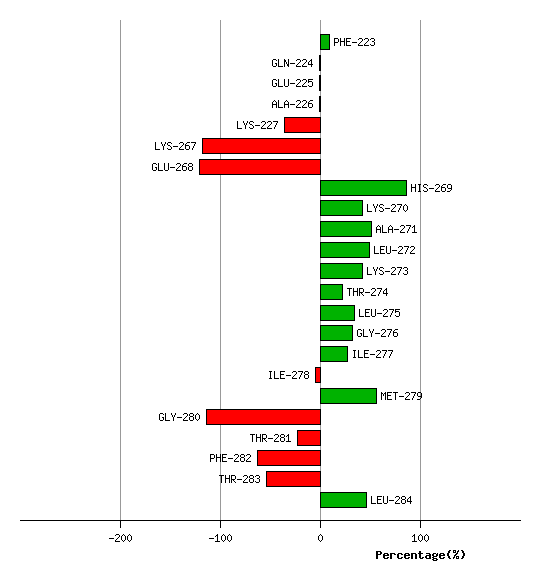
Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees